Key Result
Sclerotinia stem rot-tolerant varieties are available and efforts continue to improve varietal resistance, in both the public and private sectors. Researchers on this project have identified previously undiscovered genes associated with the plant defense response, providing valuable resources for researchers interested in developing sclerotinia-resistant canola lines.
Project Summary
This three-year study was launched to identify novel defense genes and the molecular framework underlying canola’s defense response to sclerotinia stem rot, and to answer three main biological questions:
- How does global gene expression change during whole-leaf infection in susceptible versus tolerant canola cultivars?
- What are the tissue-specific defense responses that occur in susceptible versus tolerant cultivars?
- What changes in gene expression and physiology occur in response to the biocontrol bacterium Pseudomonas chlororaphis (PA23), with and without the presence of the S. sclerotiorum pathogen?
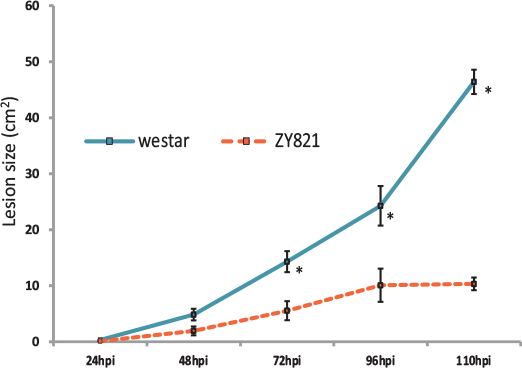
Field conditions of sclerotinia infection were accurately mimicked using a petal inoculation technique on both a tolerant B. napus genotype and a susceptible variety. Using next generation RNA sequencing and a robust bioinformatics pipeline, novel defense molecules were functionally characterized.
The team discovered over 1,200 previously undefined transcripts, including genes involved with plant defense processes and cell signaling. They also identified novel regulators of the plant defense response and found biological processes, including protein modification and non-genetic influences, which may contribute to tolerance of S. sclerotiorum.
To identify tissue-specific defense responses, three types of infected and non-infected leaf tissues were collected, 24 hours after inoculation, from both the susceptible and tolerant cultivars. RNA isolation and sequencing led to the identification and functional analysis of cell-specific genes and transcription factor regulators.
Results indicate that sclerotinia infection induces tissue-specific defense responses in susceptible leaves of B. napus plants. This data will provide a high-resolution map of the immediate plant defense response to infection.
To measure changes in response to the biocontrol bacterium PA23, B. napus plants were sprayed with solutions of PA23 alone, S. sclerotiorum ascospores only, or PA23 followed by ascospores 24 hours later. All treatments were incubated under humid conditions for 72 hours to confirm PA23’s efficacy in preventing fungal infection.
Application of PA23 reduced the number of lesions present on plant leaves by more than 91 per cent, when comparing the rate of infection to total petals in the plant canopy. Plants treated with both PA23 and S. sclerotiorum also had the fewest unique gene expression changes as a biological response, at 515 genes. This is approximately 16 times less than the 8,237 up-regulated differentially expressed genes activated in those treated with S. sclerotiorum alone.

Westar (susceptible) samples are on top and ZY821 (tolerant) samples are on the bottom. Rows of samples depict 24, 48, 72 and 96 hours post inoculation (left to right).
These findings reveal that PA23 has a significant effect on the interaction between B. napus and S. sclerotiorum, likely through modulation of defense responses in the plant and activation of unique gene expression patterns at the point of infection. This is significantly relevant for the use of PA23 as a biocontrol agent in the field and in commercial applications.
A mini review article recently published in Frontiers in Plant Science* illustrates the application of this research. With fungal pathogens capable of destroying 60 per cent of all crops in a severe epidemic, innovative plant protection solutions are an immediate priority. Integration of these cutting-edge molecular tools represents one more step for plant scientists, breeders and canola growers towards finding new and translatable solutions to protect global food systems.





