Key Result
A three-year study showed that including two resistant genes with different modes of action in clubroot resistant canola cultivars is more durable than single resistance genes against the new virulent pathogen pathotype 5X. To improve durability of resistance, growers also need a break of at least two years between canola crops.
Project Summary
Cultivar resistance is the key to managing clubroot, which continues to spread on the Canadian prairies. Recently in Alberta, 17 “new” pathogen pathotypes were identified and all appeared virulent to previous resistant cultivars in the marketplace. This erosion of single-gene resistance is also being reported across western Canada, including a new variant of pathotype 3A in Manitoba. To address this challenge, researchers investigated the efficacy and durability of canola lines carrying single and multiple clubroot resistance (CR) genes against “new” and “old” pathotypes. Researchers also assessed resistance durability under heavy (Alberta) and lighter (Saskatchewan and Manitoba) infestation situations against the predominant pathotype 3H to better understand the risk of resistance erosion in recommending a CR canola cultivar in regions with different clubroot pathogen inoculum load on the Prairies.
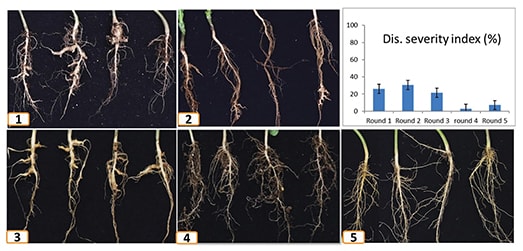
For this three-year study, 20 canola-quality B. napus inbred and hybrid lines carrying single, double and triple CR genes were produced in collaboration with Nutrien Ag Solutions. Advanced RNA sequencing methods were used to compare single- and double-gene lines to determine the modes of action of the different genes. All of the genotypes selected were resistant to the predominant pathotype 3H and other ‘old’ pathotypes (2, 5, 6, 8), however resistance to new pathotypes was unknown.
Westar and 45H29 (resistant to old pathotypes), both susceptible to the newly identified pathotype 5X, were included as controls. These lines were assessed for resistance against three field populations of the pathotype 5X using inoculum from Stephen Strelkov’s lab at the University of Alberta and under simulated intensive canola-growing conditions. The lines went through five generational cycles of exposure, with all root galls recycled back into the growth media at the end of each generation. The experiment lasted about 18 months for each pathotype and repeated once.
Researchers found that using a variety with double CR genes located on two different chromosomes of the A genome, A8 (CRB) and A3 (Rcr1 or CRM), provided moderate resistance against all 5X populations, as well as the immunity to the old pathotypes 2, 3, 5, 6 and 8. The study also showed that in response to 5X infection, many genes involved in host immunity pathways were more strongly activated in lines carrying these two CR genes, relative to those controlled by either of the single CR genes alone. Although the resistance provided by stacking two CR genes with different modes of action only increased to a moderate level, this resistance to 5X proved to be quite robust and more durable than expected. Over the five generations of exposure to the inoculum, the clubroot galls were smaller and fewer, and the inoculum levels did not increase. In some cases, inoculum levels even went down slightly.
The study also confirmed that higher inoculum loads tend to accelerate the resistance erosion, especially for canola varieties carrying only a single CR gene. Therefore, for producers with fields of heavy clubroot infestation, an extended rotation is necessary to protect the performance and durability of clubroot resistance. Previous research by Peng shows that a two-year break between canola crops can potentially reduce the pathogen inoculum in the soil by up to 90 per cent relative to one-year break. Current study results also suggest that even in areas with low levels of clubroot infestation, such as Saskatchewan and Manitoba, using CR varieties is recommended to keep inoculum levels low and delay potential outbreak. Researchers plan to see if this successful strategy of stacking two CR genes to manage 5X also applies to other new virulent pathotypes in future research.





